New IDT Multiplexed Target Amplification Technology Promises High Specificity
Integrated DNA Technologies (IDT) last week launched a multiplexed target amplification technology for next-gen sequencing, called rhAmpSeq, that it says provides high specificity and an easy workflow and promises to be cost effective.
The new amplicon solution adds to a number of other sample and library prep products that IDT, which is headquartered in Coralville, Iowa, and was acquired by Danaher last year for approximately $2 billion, has launched for the next-generation sequencing market in recent years. In 2015, for example, the firm launched an exome capture kit, which has been popular in the market, and in 2017, it partnered with Illumina to provide improved library prep multiplexing and target enrichment solutions for NGS. Later that year, IDT launched a cancer sequencing panel for acute myeloid leukemia.
In contrast to its previous target enrichment products, branded xGen, which rely on hybridization capture, the new rhAmpSeq technology uses PCR amplification and will likely compete with the AmpliSeq technology, originally developed by Life Technologies for its Ion Torrent sequencing technology and licensed by Illumina from Thermo Fisher Scientific last year for research use with Illumina’s sequencing platforms.
RhAmpSeq, which is currently only compatible with Illumina sequencing, allows users to amplify up to 5,000 targets in a single pool, with an insert size of 50 to 200 nucleotides. According to IDT’s website, it works with tissue, FFPE, or cell-free DNA samples and requires a minimum DNA input of of 10 ng.
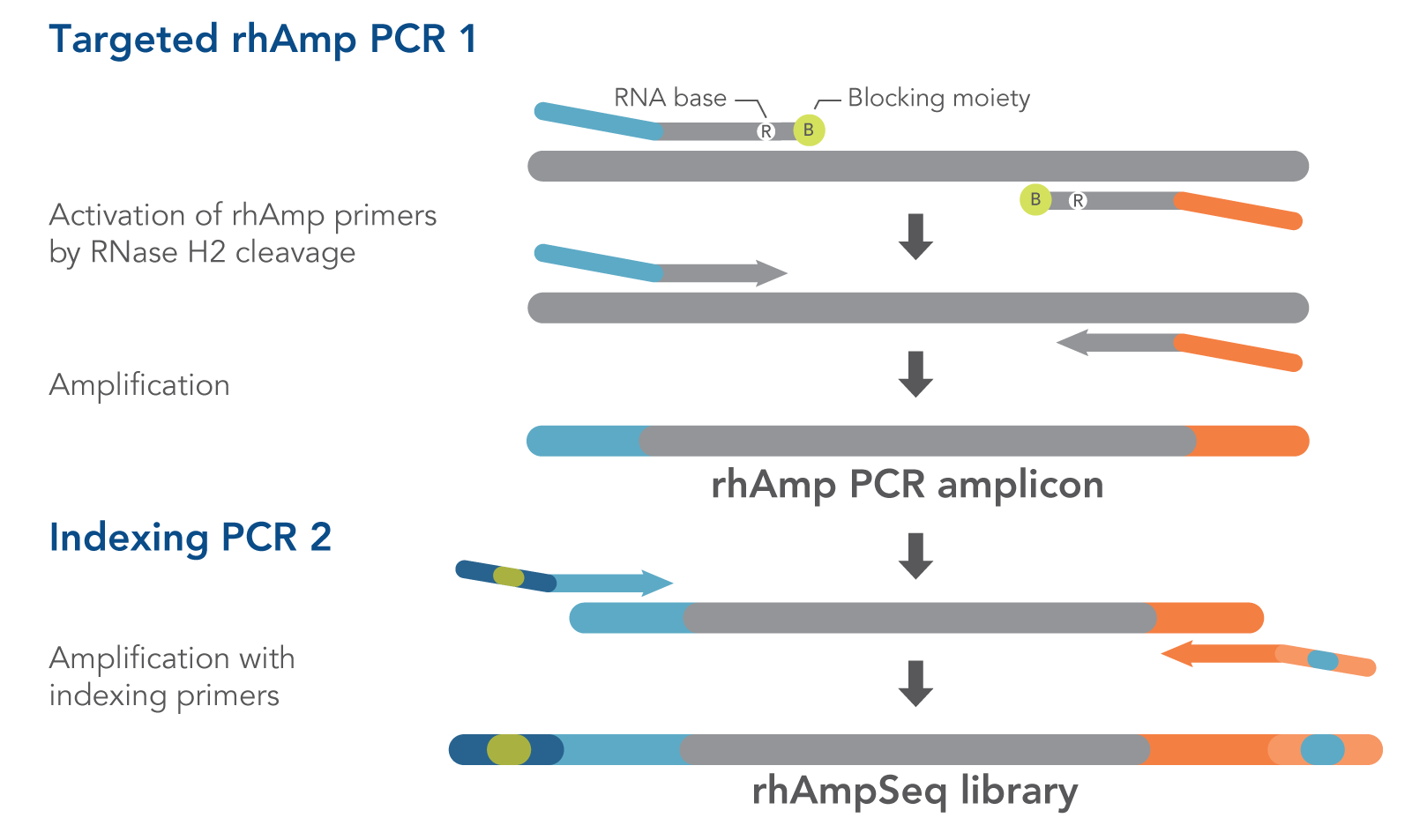
The protocol, which has two PCR amplification steps, relies on IDT’s proprietary RNase H2-dependent PCR (rhAmp PCR) technology, which the firm has already been offering for SNP genotyping.
The first PCR step uses pairs of primers that each contain an RNA base, as well as a blocking group at the 3′ end. After the primers bind to their target, RNase H2 recognizes the DNA/RNA duplex and cleaves the primer at the RNA base, thus removing the blocking group and activating the primers, so the PCR can proceed. Because RNase H2 is highly specific, it won’t activate primer dimers or primers that have bound to DNA non-specifically, which increases the specificity of the amplification step.
The second PCR uses indexing primers that also include sequencing adapters to amplify the rhAmp PCR amplicons and generate an Illumina sequencing library.
For comparison, AmpliSeq for Illumina works with as little as 1 ng input DNA, though 10 ng are recommended for most applications, according to Illumina’s website, and customers can design up to about 3,100 amplicons per pool. The AmpliSeq process involves a first PCR step, followed by the removal of primer sequences, ligation of adapters, and a second PCR step with sequencing primers.
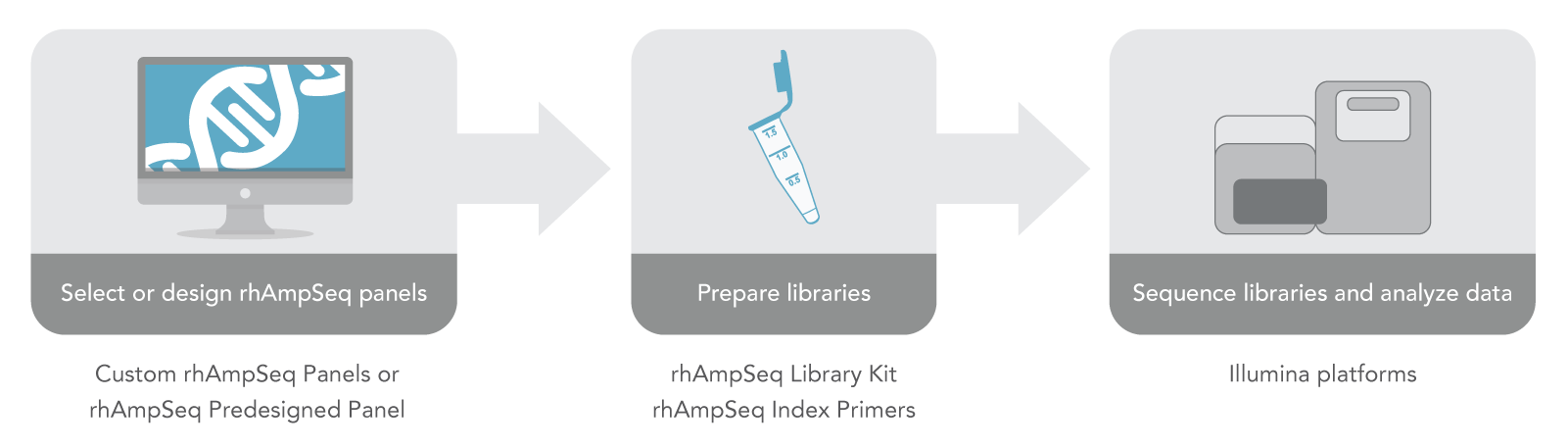
According to Allen Nguyen, director of genomics product management at IDT, compared to other vendors, which offer many pre-designed panels, IDT is focusing more on custom panels with its rhAmpSeq offering. So far, IDT provides one catalog panel, called rhAmpSeq Sample ID Panel, to help identify inadvertent swapping of human samples in a study.
Overall, rhAmpSeq will likely be useful for a number of research applications, he said, including in agricultural biotechnology for molecular breeding and trait or marker selection; CRISPR genome editing, where it can help identify off-target editing events; and human disease research.
Bruce Reisch, a professor at Cornell University’s College of Agriculture and Life Sciences, tested rhAmpSeq as an early-access customer last year as part of a multi-institutional grapevine breeding project called VitisGen2. Specifically, his team used the technology to create genetic maps of six grapevine populations, looking at the core genome they have in common, with the aim to find unique SNPs and haplotypes that would be transferable across diverse populations.
For their mapping study, which they conducted last year, they designed 2,000 amplicons, each 200 to 300 nucleotides in size, and found that more than 90 percent of them amplified in all six populations, enabling them to create maps with 1,800 to 1,900 markers each. Details of the study will be described in an upcoming scientific publication, Reisch said.
«We found the technology to be really useful,» Reisch said, adding that it has an easy workflow and very high specificity of amplification. His team has a number of other populations for which it would like to create maps, and he said he could imagine additional uses, which would depend on how rhAmpSeq is priced.
Qi Sun, co-director of the Cornell Bioinformatics Facility, who was involved in the project, said that the coverage rhAmpSeq provided across the targets was very even on the first try. «That’s very impressive,» he said, adding that there was very little off-target amplification.
Earlier during the VitisGen project, the researchers had used a different technology, genotyping by sequencing (GBS), Reisch said, but the markers they created with that «were not so easily transferable across populations.»
Editas Medicine is another early-access user of the technology. On a poster presented at the Advances in Genome Biology and Technology meeting two weeks ago, Editas researchers described how they used a 400-target rhAmpSeq panel to validate CRISPR-Cas9 off targets, comparing it to single-target amplification reactions.
They found that the single reactions required 175 times more input DNA than rhAmpSeq, which only required 40 ng of DNA, and that rhAmpSeq did not require any primer validation. Of 399 targets, only one was not covered by sequencing reads. «Multiplexing with rhAmpSeq is a powerful tool to assess specificity and will allow us to validate hundreds of potential off-target sites with nanogram amount of input DNA,» the researchers wrote.



 +502 3052-0849
+502 3052-0849 
Publicaciones recientes